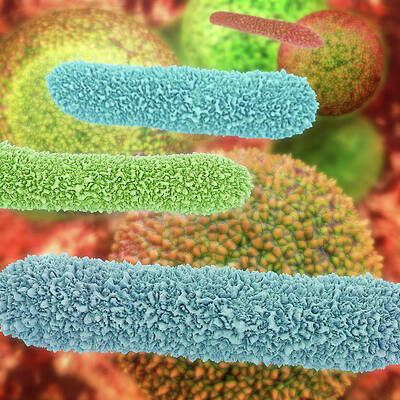
Bacteria 3d printing
Programmable microbial ink for 3D printing of living materials produced from genetically engineered protein nanofibers
Introduction
3D bioprinting technology, which is relatively well-established for printing mammalian cells in the context of tissue engineering, has more recently been applied to print microbial cells for biotechnological and biomedical applications1,2,3,4,5,6,7,8. Although inkjet printing, contact printing, screen printing, and lithographic techniques have been explored to print microbes, extrusion-based bioprinting has become one of the most widely used techniques due to its simplicity, compatibility with a variety of bioinks, and cost-effective instrumentation2,9,10,11. In an early example of this concept, a mixture of alginate and E. coli was extruded onto a printing surface consisting of calcium chloride, upon which the alginate molecules crosslink to form a solidified gel7. A similar ionic crosslinking strategy was exploited to generate photocurrent with 3D printed cyanobacteria12. In another approach, a multi-material bioink comprised of hyaluronic acid, κ-carrageenan, fumed silica, and a photo-initiator was employed to 3D-print Pseudomonas putida and Acetobacter xylinum. Also, photo-crosslinked pluronic F127 acrylate-based bioinks have been utilized to print living, responsive materials/devices, and catalytically active living materials4,6,13.
An alternative strategy made use of freeze-dried Saccharomyces cerevisiae as the primary component of a bioink formulation consisting of nanocellulose, polyethylene glycol dimethacrylate, and a photoinitiator3. The latter approach yielded remarkably high cell densities of 109 cells ml−1, but the need for freeze-drying could significantly affect the survival rate of other microbial species as well as their thixotropic behavior. In an interesting approach, the viscoelastic gel-like characteristics of Bacillus subtilis (B. subtilis) biofilms facilitated direct printing. However, the wild-type biofilms were unable to maintain the printed line widths (as they expanded three-fold in width after printing), while the engineered variants had lower storage modulus and viscosity that restricted their printing in multiple layers8. In yet another strategy, a fused deposition modeling was adapted to deposit molten agarose (75 °C) containing B. subtilis spores onto a cold substrate (16 °C), resulting in hardened patterns upon cooling5. Here, the high-temperature processing works well for spores, but limits applicability to a wide range of cell types.
However, the wild-type biofilms were unable to maintain the printed line widths (as they expanded three-fold in width after printing), while the engineered variants had lower storage modulus and viscosity that restricted their printing in multiple layers8. In yet another strategy, a fused deposition modeling was adapted to deposit molten agarose (75 °C) containing B. subtilis spores onto a cold substrate (16 °C), resulting in hardened patterns upon cooling5. Here, the high-temperature processing works well for spores, but limits applicability to a wide range of cell types.
Although the above examples demonstrate that many bioink designs have already been explored, none so far have fully leveraged the genetic programmability of microbes to rationally control the mechanical properties of the bioink. This would be advantageous for several reasons, including the possibilities of more sustainable manufacturing practices, raw material fabrication in resource-poor environments (terrestrial or extra-terrestrial), and enhanced material performance through bio-inspired design and the precision of genetic programming. In contrast to the examples described above, we envisioned to (1) design an extrudable bioink that had high print fidelity, (2) produce the bioink entirely from engineered microbes by a bottom-up approach and (3) create a programmable platform that would enable advanced functions for the macroscopic 3D living architectures, and thereby push the emerging field of living materials to unexplored frontiers3,4,5,6,7,8,9,10,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28.
In contrast to the examples described above, we envisioned to (1) design an extrudable bioink that had high print fidelity, (2) produce the bioink entirely from engineered microbes by a bottom-up approach and (3) create a programmable platform that would enable advanced functions for the macroscopic 3D living architectures, and thereby push the emerging field of living materials to unexplored frontiers3,4,5,6,7,8,9,10,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28.
In this work, we present microbial ink that is produced entirely from the genetically engineered E. coli biofilms. We show the detailed characterization of the microbial ink and demonstrate its structural and shape integrity. Further, by embedding genetically programmed E. coli cells in the microbial ink, we demonstrate the 3D printing of therapeutic living material, sequestration living material, and regulatable living material.
Results
Design strategy and production of microbial ink
There is a critical need to develop advanced bioinks with tunable mechanical strength, high cell viability, and high print fidelity11. A printable bioink requires a viscosity low enough to facilitate extrusion, but high enough to retain its shape after printing2. In this regard, shear-thinning hydrogels, which decrease their viscosity with increasing shear stress, are an attractive option. Moreover, it should be noted that bioinks are biocompatible materials typically meant to recapitulate an extracellular matrix (ECM) to provide a congenial environment for the growth of living cells with predefined structures and functions. We envisioned that instead of embedding microbes in an ECM-mimicking bioink, we could repurpose the ECM of the microbial biofilm itself to serve as a programmable bioink. This idea is built on our earlier work, wherein we showed that the native proteinaceous curli nanofibers of an E. coli biofilm ECM can be genetically engineered by fusing functional peptides/proteins to the curli CsgA monomer to produce a shear-thinning hydrogel18. However, in order to create a bioink with the desired viscoelastic performance, we introduced a genetically programmed crosslinking strategy, inspired by fibrin (Fig.
A printable bioink requires a viscosity low enough to facilitate extrusion, but high enough to retain its shape after printing2. In this regard, shear-thinning hydrogels, which decrease their viscosity with increasing shear stress, are an attractive option. Moreover, it should be noted that bioinks are biocompatible materials typically meant to recapitulate an extracellular matrix (ECM) to provide a congenial environment for the growth of living cells with predefined structures and functions. We envisioned that instead of embedding microbes in an ECM-mimicking bioink, we could repurpose the ECM of the microbial biofilm itself to serve as a programmable bioink. This idea is built on our earlier work, wherein we showed that the native proteinaceous curli nanofibers of an E. coli biofilm ECM can be genetically engineered by fusing functional peptides/proteins to the curli CsgA monomer to produce a shear-thinning hydrogel18. However, in order to create a bioink with the desired viscoelastic performance, we introduced a genetically programmed crosslinking strategy, inspired by fibrin (Fig. 1)29. Fibrin is a protein involved in the clotting cascade, which activates its polymerization to form blood clots. Fibrin’s polymerization is driven in part by noncovalent interactions between an alpha-chain domain present on the N-terminus of one fibrin monomer (i.e., the “knob” domain) and a gamma-chain domain on the C-terminus (i.e., the “hole” domain) of an adjacent monomer29. Our microbial ink design repurposes this binding interaction between alpha and gamma modules, i.e., the knob-hole interaction, to introduce non-covalent crosslinks between nanofibers and enhance mechanical robustness while maintaining shear-thinning properties (Fig. 1). It should also be noted that fibers formed from the self-assembly of CsgA are highly stable and resistant to proteolytic, detergent-induced, and heat-based denaturation30.
1)29. Fibrin is a protein involved in the clotting cascade, which activates its polymerization to form blood clots. Fibrin’s polymerization is driven in part by noncovalent interactions between an alpha-chain domain present on the N-terminus of one fibrin monomer (i.e., the “knob” domain) and a gamma-chain domain on the C-terminus (i.e., the “hole” domain) of an adjacent monomer29. Our microbial ink design repurposes this binding interaction between alpha and gamma modules, i.e., the knob-hole interaction, to introduce non-covalent crosslinks between nanofibers and enhance mechanical robustness while maintaining shear-thinning properties (Fig. 1). It should also be noted that fibers formed from the self-assembly of CsgA are highly stable and resistant to proteolytic, detergent-induced, and heat-based denaturation30.
a E. coli was genetically engineered to produce microbial ink by fusing α (knob) and γ (hole) protein domains, derived from fibrin to the main structural component of curli nanofibers, CsgA. Upon secretion, the CsgA-α and CsgA-γ monomers self-assemble into nanofibers crosslinked by the knob-hole binding interaction. b The knob and hole domains are derived from fibrin, where they play a key role in supramolecular polymerization during blood clot formation. c The protocol to produce microbial ink from the engineered protein nanofibers involves standard bacterial culture, limited processing steps, and no addition of exogenous polymers. Microbial ink was 3D printed to obtain functional living materials.
coli was genetically engineered to produce microbial ink by fusing α (knob) and γ (hole) protein domains, derived from fibrin to the main structural component of curli nanofibers, CsgA. Upon secretion, the CsgA-α and CsgA-γ monomers self-assemble into nanofibers crosslinked by the knob-hole binding interaction. b The knob and hole domains are derived from fibrin, where they play a key role in supramolecular polymerization during blood clot formation. c The protocol to produce microbial ink from the engineered protein nanofibers involves standard bacterial culture, limited processing steps, and no addition of exogenous polymers. Microbial ink was 3D printed to obtain functional living materials.
Full size image
Using the Biofilm Integrated Nanofiber Display (BIND) technology developed in our laboratory25, we genetically grafted the “knob” and “hole” protein domains to the N- and C-terminus of CsgA, respectively, to create the fusion proteins CsgA-α and CsgA-γ (Fig. 1, Supplementary Table 1). CsgA-α and CsgA-γ were expressed separately in engineered E. coli strain PQN4 along with the auxiliary curli genes necessary for secretion and assembly, and the resulting curli nanofibers were imaged using transmission electron microscopy (TEM). After staining with 1% uranyl formate, the nanofibers showed diameters of ~5.5 nm (CsgA-α) and ~6.7 nm (CsgA-γ) (Fig. 2a). Notably, curli nanofibers composed of wild-type CsgA have diameters of ~4 nm25. Thus, the observed trend in nanofiber diameters is qualitatively consistent with the relative sizes of the fused domains – 11 amino acids for “knob” (CsgA-α) and 127 amino acids for “hole” (CsgA-γ). When the two types of E. coli cells, each expressing either CsgA-α or CsgA-γ, were co-cultured (CsgA-αγ), they produced nanofibers that display “knob” and “hole” domains. TEM imaging showed three nanofiber populations with diameters of ~5.
1, Supplementary Table 1). CsgA-α and CsgA-γ were expressed separately in engineered E. coli strain PQN4 along with the auxiliary curli genes necessary for secretion and assembly, and the resulting curli nanofibers were imaged using transmission electron microscopy (TEM). After staining with 1% uranyl formate, the nanofibers showed diameters of ~5.5 nm (CsgA-α) and ~6.7 nm (CsgA-γ) (Fig. 2a). Notably, curli nanofibers composed of wild-type CsgA have diameters of ~4 nm25. Thus, the observed trend in nanofiber diameters is qualitatively consistent with the relative sizes of the fused domains – 11 amino acids for “knob” (CsgA-α) and 127 amino acids for “hole” (CsgA-γ). When the two types of E. coli cells, each expressing either CsgA-α or CsgA-γ, were co-cultured (CsgA-αγ), they produced nanofibers that display “knob” and “hole” domains. TEM imaging showed three nanofiber populations with diameters of ~5. 5, ~6.7, and ~10 nm (Fig. 2a, Supplementary Fig. 1). We attribute the 10 nm diameter nanofibers to supramolecular crosslinking mediated by noncovalent interactions of the “knob” and “hole” domains (Fig. 1). We then created hydrogels from the microbial cultures using a simple filtration protocol, as described in our earlier reports18. Briefly, the microbial culture was filtered through a nylon membrane to concentrate the curli nanofibers, and then treated with guanidinium chloride, nuclease, and sodium dodecyl sulfate to obtain cell-free hydrogels composed of the designed curli nanofibers (Fig. 2b)18. Field-emission scanning electron microscopy (FESEM) indicated a fibrous microstructure for all three hydrogels (CsgA-α, CsgA-γ and the co-culture CsgA-αγ), with the fiber alignment suggesting hierarchical assembly through the lateral association of functional curli nanofibers (Fig. 2b).
5, ~6.7, and ~10 nm (Fig. 2a, Supplementary Fig. 1). We attribute the 10 nm diameter nanofibers to supramolecular crosslinking mediated by noncovalent interactions of the “knob” and “hole” domains (Fig. 1). We then created hydrogels from the microbial cultures using a simple filtration protocol, as described in our earlier reports18. Briefly, the microbial culture was filtered through a nylon membrane to concentrate the curli nanofibers, and then treated with guanidinium chloride, nuclease, and sodium dodecyl sulfate to obtain cell-free hydrogels composed of the designed curli nanofibers (Fig. 2b)18. Field-emission scanning electron microscopy (FESEM) indicated a fibrous microstructure for all three hydrogels (CsgA-α, CsgA-γ and the co-culture CsgA-αγ), with the fiber alignment suggesting hierarchical assembly through the lateral association of functional curli nanofibers (Fig. 2b).
a Transmission electron microscope (TEM) images of self-assembled nanofibers of CsgA-α, CsgA-γ and CsgA-αγ (co-culture of CsgA-α and CsgA-γ) after recombinant expression. Representative images from three independent samples were reported. b Optical images of CsgA-α, CsgA-γ and microbial ink CsgA-αγ hydrogels with the corresponding field-emission scanning electron microscope (FESEM) images show the presence of aligned microscopic fiber bundles. Representative images from three independent samples were reported.
Full size image
Characterization and 3D printing of microbial ink
We investigated the rheological properties of CsgA-α, CsgA-γ, and CsgA-αγ hydrogels, to validate their potential as extrudable bioinks. Frequency sweep experiments revealed that the storage modulus (G’) of the CsgA-αγ hydrogel was several-fold higher than that of the CsgA-α and CsgA-γ hydrogels alone, while the G’ of all the hydrogels were higher than their loss modulus (G”) by an order of magnitude (Fig. 3a). Strain sweep experiments showed that the hydrogels were stable up to ~10% strain, above which a crossover point is observed as G’ decreased and G” increased. (Fig. 3b). The viscosity of all the hydrogels was also found to decrease with increasing shear rate, which indicates their shear-thinning behavior (Fig. 3c). Similarly, the shear modulus (G) of CsgA-αγ was higher than that of CsgA-α and CsgA-γ by 6- and 3-fold, respectively (Fig. 3d). The yield stress (σy) of CsgA-αγ was nearly twice that of CsgA-α and CsgA-γ (Fig. 3e). From all the above experiments, it is clear that supramolecular crosslinking of “knob” and “hole” domains in CsgA-αγ significantly increased the G’, G, σy, and viscosity, making it better suited than CsgA-α or CsgA-γ for extrusion printing2,11. On the other hand, when CsgA-α and CsgA-γ fibers expressed in separate cultures were mixed (CsgA-αγ-mix) in a 1:1 volume ratio also yielded hydrogels with rheological properties similar to CsgA-αγ, which further confirms our hypothesis about supramolecular crosslinking between the complementary fibers (Supplementary Figs.
3a). Strain sweep experiments showed that the hydrogels were stable up to ~10% strain, above which a crossover point is observed as G’ decreased and G” increased. (Fig. 3b). The viscosity of all the hydrogels was also found to decrease with increasing shear rate, which indicates their shear-thinning behavior (Fig. 3c). Similarly, the shear modulus (G) of CsgA-αγ was higher than that of CsgA-α and CsgA-γ by 6- and 3-fold, respectively (Fig. 3d). The yield stress (σy) of CsgA-αγ was nearly twice that of CsgA-α and CsgA-γ (Fig. 3e). From all the above experiments, it is clear that supramolecular crosslinking of “knob” and “hole” domains in CsgA-αγ significantly increased the G’, G, σy, and viscosity, making it better suited than CsgA-α or CsgA-γ for extrusion printing2,11. On the other hand, when CsgA-α and CsgA-γ fibers expressed in separate cultures were mixed (CsgA-αγ-mix) in a 1:1 volume ratio also yielded hydrogels with rheological properties similar to CsgA-αγ, which further confirms our hypothesis about supramolecular crosslinking between the complementary fibers (Supplementary Figs. 2–3).
2–3).
The storage modulus (G’) and loss modulus (G”) under frequency sweep (a) and oscillatory sweep (b). c Viscosity as a function of shear rate, d Shear modulus (n = 3) **p = 0.0014, ***p = 0.0004 and e yield stress (n = 3) **p = 0.0061, **p = 0.003, one-way ANOVA followed by Dunnett’s test. f Printed line diameter as a function of feed rates ranging from 2 to 10 mm s−1 at 20 psi pressure (n > 10). g Images of filament collapse test and h. the plot of deflection angle versus pillar gap distances (n = 3). Experimental data: solid line, theoretically predicted data: dotted line. Data represented as mean ± standard deviation. **p ≤ 0.01, ***p ≤ 0.001, one-way ANOVA followed by Dunnett’s test. 3D printed structures using the microbial ink CsgA-αγ i single-layer grid, j 10-layer square, k 10-layer circle, and l 21-layer solid cone. Insets in (j–l). are corresponding top views. Scale bar 1 mm.
Insets in (j–l). are corresponding top views. Scale bar 1 mm.
Full size image
We then tested the printability of the CsgA-α, CsgA-γ, and CsgA-αγ hydrogel-based bioinks using a customized 3D printer (Supplementary Fig. 4). First, the hydrogel-based bioinks were extruded under a range of feed rates (2–10 mm s−1) and pressures (20–40 psi) to understand their printing performance (Fig. 3f, Supplementary Figs. 5−10). The printed line widths of CsgA-α and CsgA-γ bioinks were nearly two times that of CsgA-αγ for the same feed rate and pressure, indicating the superior structural integrity of CsgA-αγ (Fig. 3f, Supplementary Figs. 5–10). Subsequently, we tested the shape fidelity of the bioinks according to a published protocol (filament collapse test) to provide a quantitative comparison between bioinks31. A single line (filament) of each bioink was extruded at a nozzle moving speed of 5 mm s−1 on a platform with pillars at known gap distances, to bridge the pillar gaps (Fig. 3g, Supplementary Movie 1). The CsgA-α and CsgA-γ bioinks were unable to bridge gap distances of 8 mm and above, whereas the CsgA-αγ bioink was able to support its own weight for gap distances as large as 16 mm. This remarkable property, along with the higher σy and viscosity, suggests that the supramolecular knob-hole crosslinks allow for fast reassembly after extrusion—an important feature for optimal bioink performance32,33. Shape fidelity was assessed quantitatively by measuring the angles of deflection of the overhung bioink fibers under gravitational force (Fig. 3h)31. When the angles of deflection are plotted against the half gap distances, the resulting slope will decrease for the bioink with higher σy31. This experimental data was also consistent with the reported theoretical model, while the deviation of experimental and predicted slopes was also in line with the original report, which observed that the model overestimated the angles of deflection, likely due to the exclusion of gel viscoelasticity and surface tension from the theoretical model31.
3g, Supplementary Movie 1). The CsgA-α and CsgA-γ bioinks were unable to bridge gap distances of 8 mm and above, whereas the CsgA-αγ bioink was able to support its own weight for gap distances as large as 16 mm. This remarkable property, along with the higher σy and viscosity, suggests that the supramolecular knob-hole crosslinks allow for fast reassembly after extrusion—an important feature for optimal bioink performance32,33. Shape fidelity was assessed quantitatively by measuring the angles of deflection of the overhung bioink fibers under gravitational force (Fig. 3h)31. When the angles of deflection are plotted against the half gap distances, the resulting slope will decrease for the bioink with higher σy31. This experimental data was also consistent with the reported theoretical model, while the deviation of experimental and predicted slopes was also in line with the original report, which observed that the model overestimated the angles of deflection, likely due to the exclusion of gel viscoelasticity and surface tension from the theoretical model31. We then utilized the CsgA-αγ bioink to 3D-print defined patterns and shapes. A single-layer grid shows the finer line structures of the printed pattern obtained with a resolution of ~300 μm from a 27 G needle (Fig. 3i). The multilayered architectures presented in Fig. 3j (10-layered square), Fig. 3k (10-layered circle) and Fig. 3l (21-layered solid cone) demonstrated the structural integrity of the microbial ink.
We then utilized the CsgA-αγ bioink to 3D-print defined patterns and shapes. A single-layer grid shows the finer line structures of the printed pattern obtained with a resolution of ~300 μm from a 27 G needle (Fig. 3i). The multilayered architectures presented in Fig. 3j (10-layered square), Fig. 3k (10-layered circle) and Fig. 3l (21-layered solid cone) demonstrated the structural integrity of the microbial ink.
3D printing of functional living materials
After demonstrating the printing performance of the microbial ink, we introduced the genetically engineered microbes to the hydrogel to produce 3D-printed living functional architectures. Herein, we present a living material for therapeutic applications, wherein a chemical inducer, isopropyl β-D-1-thiogalactopyranoside (IPTG) was utilized to signal the programmed E. coli (PQN4-Azu) to synthesize on demand an anticancer biologic drug, azurin, and secrete it into the extracellular milieu (Fig. 4a, Supplementary Fig. 11)34. The microbial ink, laden with PQN4-Azu cells, was used to print a 2D capsule pattern that was incubated in the lysogeny broth (LB) media with (+) and without (−) the inducer IPTG. After 24 and 48 h of incubation, the detection of the secreted azurin by an anti-azurin antibody illustrated the functioning of the 3D-printed therapeutic living material (Fig. 4a). Next, we produced a living material designed to sequester a toxic chemical, Bisphenol A (BPA). For this demonstration, we grafted a BPA-binding peptide domain to CsgA (CsgA-BPABP) and loaded the PQN4 cells expressing CsgA-BPABP into the microbial ink (Fig. 4b, Supplementary Fig. 12)35. After printing a 2D pattern with the cell-laden ink, the pattern was incubated in LB media with 1 mM BPA. Liquid chromatography-mass spectroscopy (LC-MS) analysis showed that the microbial ink embedded with CsgA-BPA biofilm sequestered nearly 8% and 27% of the BPA after 12 and 24 h of incubation, respectively, while a negative control pattern made with microbial ink only, showed no appreciable BPA sequestration (Fig.
11)34. The microbial ink, laden with PQN4-Azu cells, was used to print a 2D capsule pattern that was incubated in the lysogeny broth (LB) media with (+) and without (−) the inducer IPTG. After 24 and 48 h of incubation, the detection of the secreted azurin by an anti-azurin antibody illustrated the functioning of the 3D-printed therapeutic living material (Fig. 4a). Next, we produced a living material designed to sequester a toxic chemical, Bisphenol A (BPA). For this demonstration, we grafted a BPA-binding peptide domain to CsgA (CsgA-BPABP) and loaded the PQN4 cells expressing CsgA-BPABP into the microbial ink (Fig. 4b, Supplementary Fig. 12)35. After printing a 2D pattern with the cell-laden ink, the pattern was incubated in LB media with 1 mM BPA. Liquid chromatography-mass spectroscopy (LC-MS) analysis showed that the microbial ink embedded with CsgA-BPA biofilm sequestered nearly 8% and 27% of the BPA after 12 and 24 h of incubation, respectively, while a negative control pattern made with microbial ink only, showed no appreciable BPA sequestration (Fig. 4b). Finally, we show that the cell growth within the printed material can be regulated by inducing a genetic circuit (Fig. 4c, Supplementary Fig. 13). To accomplish this, E. coli (PQN4-MazF) cells were programmed to express (upon induction with IPTG) the endoribonuclease toxin, MazF, that inhibits protein synthesis by cleaving mRNA, and can arrest cell growth and/or lead to cell death36. PQN4-MazF cells in the printed structure were found to proliferate in the absence of IPTG, but after 2 h of IPTG induction, the colony-forming unit (CFU) count reduced by nearly two orders of magnitude due to the expression of MazF (Fig. 4c). However, subsequently, the cell growth was restored to some extent, likely due to the native MazFE toxin-antitoxin system in E. coli (Fig. 4c)36. Such a regulation system can be further engineered to effectively control the cell growth and/or to induce cell death depending on the need.
4b). Finally, we show that the cell growth within the printed material can be regulated by inducing a genetic circuit (Fig. 4c, Supplementary Fig. 13). To accomplish this, E. coli (PQN4-MazF) cells were programmed to express (upon induction with IPTG) the endoribonuclease toxin, MazF, that inhibits protein synthesis by cleaving mRNA, and can arrest cell growth and/or lead to cell death36. PQN4-MazF cells in the printed structure were found to proliferate in the absence of IPTG, but after 2 h of IPTG induction, the colony-forming unit (CFU) count reduced by nearly two orders of magnitude due to the expression of MazF (Fig. 4c). However, subsequently, the cell growth was restored to some extent, likely due to the native MazFE toxin-antitoxin system in E. coli (Fig. 4c)36. Such a regulation system can be further engineered to effectively control the cell growth and/or to induce cell death depending on the need.
 4: 3D printing of functional living materials.
4: 3D printing of functional living materials.a Genetic design of E. coli (PQN4-Azu) cells, programmed to secrete an anticancer biologic drug azurin along with image of printed living material (top), and the incorporation of PQN4-Azu cells into the CsgA-αγ microbial ink (middle). Western blot (bottom) shows the difference in azurin detected in the supernatant of the printed structure with and without IPTG induction. b Genetic design of E. coli (PQN4-BPA), programmed to produce extracellular fibers displaying BPA-binding peptide (CsgA-BPABP) along with image of printed living material (top), and incorporation of CsgA-BPABP biofilm into microbial ink (middle). After 12 and 24 h, the BPA concentration (bottom) in the supernatant of the printed structure was analyzed by LCMS. Dotted line represents the initial BPA concentration of 1 mM. n = 3. c Genetic design of E. coli (PQN4-MazF) cells, programmed to express an endoribonuclease MazF, that inhibits/arrests cell growth along with image of printed living material (top), and incorporation of PQN4-MazF cells into microbial ink (middle). CFU count from printed structure over time, with and without IPTG induction (bottom). n = 3. Scale bar 5 mm. Data represented as mean ± standard deviation.
CFU count from printed structure over time, with and without IPTG induction (bottom). n = 3. Scale bar 5 mm. Data represented as mean ± standard deviation.
Full size image
In summary, we have genetically engineered the ECM of E. coli biofilms to produce a shear-thinning hydrogel by supramolecular crosslinking of fibrin-inspired recombinant protein nanofibers. Instead of using an external biocompatible material as the bioink, we have shown that a cell-laden bioink with target rheological and functional properties can be created purely through genetic engineering and minimal processing. The printability, structural integrity, and print fidelity of the microbial ink was demonstrated with the aid of detailed rheological studies and filament collapse tests. By incorporating programmed microbes/biofilms into the microbial ink, we have 3D-printed living materials that can be chemically induced to release the anticancer drug azurin, remove BPA from their surroundings, and regulate their own cell growth. The microbial ink design can be further customized for various biotechnological and biomedical applications using the ever-growing toolkit of biological parts being developed by synthetic biologists. Especially if combined with other materials technologies, such as those that are already incorporating living cells into structural building materials37, our microbial bioink could also be particularly useful for structure building in space or extraterrestrial habitats, where raw material transport is difficult, making on-demand generation of building materials from very limited resources essential37.
The microbial ink design can be further customized for various biotechnological and biomedical applications using the ever-growing toolkit of biological parts being developed by synthetic biologists. Especially if combined with other materials technologies, such as those that are already incorporating living cells into structural building materials37, our microbial bioink could also be particularly useful for structure building in space or extraterrestrial habitats, where raw material transport is difficult, making on-demand generation of building materials from very limited resources essential37.
Methods
Plasmids for production of microbial ink
The CsgA-α construct was designed by fusing the fibrin-derived alpha (knob) peptide to the N-terminus of CsgA with an intervening 12-amino-acid flexible linker. However, in case of CsgA-γ construct, the gene encoding the fibrin-derived gamma (hole) protein was fused to the C-terminus of CsgA with an intervening 36-amino-acid flexible linker. Both genes encoding CsgA-α and CsgA-γ were synthesized (Integrated DNA Technologies) and cloned into pET21d vector using isothermal Gibson assembly (New England Biolabs). Both pET21dCsgA-α and pET21dCsgA-γ plasmids were transformed into PQN4, an E. coli cell strain derived from LSR10 (MC4100, ΔcsgA, λ(DE3), CamR) with the deletion of curli operon (∆csgBACEFG). Both pET21dCsgA-α and pET21dCsgA-γ plasmids contained curli operon genes co-transcribed with the engineered csgA, i.e., csgC, csgE, csgF, and csgG, which encodes the proteins necessary for the biosynthesis of curli fibers. In both these plasmids, the csgB gene was deleted from the curli operon in order to secrete CsgA fused alpha or gamma proteins and self-assemble them to functional curli fibers (CsgA-α or CsgA-γ) in the culture medium, without anchoring (ΔcsgB) to the bacterial surface.
Both genes encoding CsgA-α and CsgA-γ were synthesized (Integrated DNA Technologies) and cloned into pET21d vector using isothermal Gibson assembly (New England Biolabs). Both pET21dCsgA-α and pET21dCsgA-γ plasmids were transformed into PQN4, an E. coli cell strain derived from LSR10 (MC4100, ΔcsgA, λ(DE3), CamR) with the deletion of curli operon (∆csgBACEFG). Both pET21dCsgA-α and pET21dCsgA-γ plasmids contained curli operon genes co-transcribed with the engineered csgA, i.e., csgC, csgE, csgF, and csgG, which encodes the proteins necessary for the biosynthesis of curli fibers. In both these plasmids, the csgB gene was deleted from the curli operon in order to secrete CsgA fused alpha or gamma proteins and self-assemble them to functional curli fibers (CsgA-α or CsgA-γ) in the culture medium, without anchoring (ΔcsgB) to the bacterial surface.
Plasmids used to make functional microbial ink
The pET21dAzu plasmid was created similar to that of pET21dCsgA-α by replacing the gene of csgA-α with that of azurin, while we retained the SEC (N-terminal signal sequence) and N22 (N-terminal curli-specific targeting sequence) to allow secretion of azurin into the extracellular milieu. In case of pET21dCsgA-BPA plasmid (similar to pET21dCsgA-γ), the gene encoding BPA binding peptide was fused to the C-terminus of CsgA via a 36-amino-acid flexible linker. The MazF plasmid pL-MazF was derived from IPTG inducible plasmid pL6FO, and contains LacI repressor and kanamycin resistance genes, with a pLac promoter upstream of a mazF sequence derived from the genome of E. coli K-12 MG1655 (EcoCyc Accession EG11249)38. All plasmids used for creating functional microbial inks were transformed into E. coli strain PQN4.
In case of pET21dCsgA-BPA plasmid (similar to pET21dCsgA-γ), the gene encoding BPA binding peptide was fused to the C-terminus of CsgA via a 36-amino-acid flexible linker. The MazF plasmid pL-MazF was derived from IPTG inducible plasmid pL6FO, and contains LacI repressor and kanamycin resistance genes, with a pLac promoter upstream of a mazF sequence derived from the genome of E. coli K-12 MG1655 (EcoCyc Accession EG11249)38. All plasmids used for creating functional microbial inks were transformed into E. coli strain PQN4.
Microbial production of engineered nanofibers
pET21dCsgA-α and pET21dCsgA-γ plasmids were transformed into PQN4 cells and streaked onto lysogeny broth (LB) agar plates containing 100 µg ml−1 carbenicillin and 0.5% glucose (mv−1) for catabolite repression of T7RNAP and incubated overnight at 37 °C. One colony was picked from each plate of PQN4CsgA-α and PQN4CsgA-γ, and cultured separately at 37 °C in 5 ml LB media, 100 µg ml−1 carbenicillin and 2% glucose (mv−1). The overnight cultures of PQN4CsgA-α and PQN4CsgA-γ were transferred to a fresh 500 ml LB media containing 100 µg ml−1 carbenicillin and cultured upon IPTG induction either separately or together. This mono- or co-culture of PQN4CsgA-α and PQN4CsgA-γ was placed in incubator shakers (225 rpm, 37 °C) for 48 h to express the engineered proteins CsgA-α and CsgA-γ, and self-assemble them into functional curli nanofibers.
The overnight cultures of PQN4CsgA-α and PQN4CsgA-γ were transferred to a fresh 500 ml LB media containing 100 µg ml−1 carbenicillin and cultured upon IPTG induction either separately or together. This mono- or co-culture of PQN4CsgA-α and PQN4CsgA-γ was placed in incubator shakers (225 rpm, 37 °C) for 48 h to express the engineered proteins CsgA-α and CsgA-γ, and self-assemble them into functional curli nanofibers.
Preparation of CsgA-
α, CsgA-γ and CsgA-αγ hydrogels (microbial ink)The 48 h bacterial culture (500 ml) with CsgA-α, CsgA-γ or CsgA-αγ nanofibers was treated with 0.8 M (final concentration) guanidinium chloride (GdmCl) and stored at 4 °C for 1 h. Subsequently, the engineered nanofibers were concentrated on a polycarbonate membrane with 11 μm pores (EMD Millipore) using vacuum filtration. The engineered nanofibers deposited on filter membrane were treated with 8 M GdmCl for 5 min. After which, it was vacuum filtered and washed with sterile deionized (DI) water (50 ml) twice to remove the lysed bacterial debris. Then, the nucleic acids bound to the curli fibers were removed by 10 min incubation with nuclease solution (Benzonase, Sigma-Aldrich, 1.5 U ml−1), which was followed by water washes (50 ml twice). Finally, the engineered curli nanofibers present on the filter membrane were incubated with 5% (mv−1 in water) sodium dodecyl sulfate (gelator) for 5 min, followed by vacuum filtration and DI water washes (50 ml twice). The hydrogel of functional curli nanofibers (CsgA-α, CsgA-γ or CsgA-αγ) formed on the filter membrane was scraped off and stored at 4 °C.
After which, it was vacuum filtered and washed with sterile deionized (DI) water (50 ml) twice to remove the lysed bacterial debris. Then, the nucleic acids bound to the curli fibers were removed by 10 min incubation with nuclease solution (Benzonase, Sigma-Aldrich, 1.5 U ml−1), which was followed by water washes (50 ml twice). Finally, the engineered curli nanofibers present on the filter membrane were incubated with 5% (mv−1 in water) sodium dodecyl sulfate (gelator) for 5 min, followed by vacuum filtration and DI water washes (50 ml twice). The hydrogel of functional curli nanofibers (CsgA-α, CsgA-γ or CsgA-αγ) formed on the filter membrane was scraped off and stored at 4 °C.
Field-emission scanning electron microscopy (FESEM) sample preparation and imaging
FESEM samples were prepared by fixing with 2% (w v−1) glutaraldehyde and 2% (w v−1) paraformaldehyde at room temperature, overnight. The samples were gently washed with water, and the solvent was gradually exchanged to ethanol with an increasing ethanol 15-min incubation step gradient (25, 50, 75, and 100% (v v−1) ethanol). The samples were then dried in a critical point dryer, placed onto SEM sample holders using silver adhesive (Electron Microscopy Sciences), and sputtered until they were coated in a 10–20 nm layer of Pt/Pd. Images were acquired using a Zeiss Ultra55 FESEM equipped with a field emission gun operating at 5–10 kV. Representative images from three independent samples were reported.
The samples were then dried in a critical point dryer, placed onto SEM sample holders using silver adhesive (Electron Microscopy Sciences), and sputtered until they were coated in a 10–20 nm layer of Pt/Pd. Images were acquired using a Zeiss Ultra55 FESEM equipped with a field emission gun operating at 5–10 kV. Representative images from three independent samples were reported.
TEM sample preparation and imaging
Ten microliters of the cultures of CsgA-α, CsgA-γ or CsgA-αγ was drop-casted onto formvar-carbon grids (Electron Microscopy Sciences), washed with DI water (thrice), and stained with 1% uranyl formate before analysis on a JEOL 1200 TEM. Representative images from three independent samples were reported.
Optical Images
Optical images were acquired using a Canon EOS Rebel SL3 Digital SLR Camera equipped with XIT 58 mm 0.43 Wide Angle Lens and XIT 58 mm 2.2x Telephoto Lens. Representative images from three independent samples were reported.
Rheology studies of the hydrogels
The viscoelastic properties of the hydrogels were determined using a Discovery Hybrid Rheometer-3 and TRIOS software (TA Instruments, New Castle, DE). Hydrogels were loaded between a Peltier plate and a 20 mm plate geometry. The excess sample was trimmed along the edge of the 20 mm plate. Samples were surrounded with mineral oil to prevent dehydration. Time sweep experiments were conducted under continuous oscillations at 0.1 Hz with an imposed shear strain of 0.5%. After the storage modulus (G’) reached a plateau, a frequency sweep routine was applied under an oscillatory shear strain of 0.5% with the frequency increasing from 0.01 to 10 Hz. The storage modulus (G’) and loss modulus (G”) were recorded for both time and frequency sweep. The plateau storage modulus in the time sweep experiment was taken as the linear shear modulus. To probe the strain dependence of viscoelastic properties, samples were subjected to a strain sweep routine at a frequency of 0. 1 Hz with the oscillatory strain increasing from 0.01 to 100%. To measure the viscosity of samples, samples were loaded between a Peltier plate and a 20 mm plate and a flow sweep routine was performed with the shear rate increasing from 0.01 to 100 Hz. To determine the yield stress of samples, samples were subjected to a stress sweep routine at a frequency of 1 Hz with the oscillatory stress increasing from 1 to 200 Pa. The yield stress is defined as the oscillatory stress where the storage modulus decreases to 50% smaller than the linear shear modulus. Data obtained from at least three independent samples were reported.
1 Hz with the oscillatory strain increasing from 0.01 to 100%. To measure the viscosity of samples, samples were loaded between a Peltier plate and a 20 mm plate and a flow sweep routine was performed with the shear rate increasing from 0.01 to 100 Hz. To determine the yield stress of samples, samples were subjected to a stress sweep routine at a frequency of 1 Hz with the oscillatory stress increasing from 1 to 200 Pa. The yield stress is defined as the oscillatory stress where the storage modulus decreases to 50% smaller than the linear shear modulus. Data obtained from at least three independent samples were reported.
3D Printing of the hydrogels (microbial ink)
Prior to printing, the bioinks were transferred into a 10-ml Luer-Lok™ syringe and centrifuged at 112 × g for 2 min to remove any air bubbles. The standard needle used was a 27 G tip with a premade ¼” blunt end from Fisnar. Bioprinting was performed using an ANET A8 (Shenzhen Anet Technology Co) 3D printer that was upgraded into an extrusion bioprinter. The bioprinting was first conducted at a range of feed rates (2–10 mm s−1) and pressures (20–40 psi) to understand their effects on the extrusion of the bioinks. For bioprinting of actual patterns, the feed rate was kept consistent at 2.5 mm s−1, with a constant pressure of ~20 psi. The patterns were designed using Solidworks 3D design software (Dassault Systèmes SE). The 3D STL files were sliced using the slic3r engine from Repetier Host, which served as the main program for operating the extrusion bioprinter39.
The bioprinting was first conducted at a range of feed rates (2–10 mm s−1) and pressures (20–40 psi) to understand their effects on the extrusion of the bioinks. For bioprinting of actual patterns, the feed rate was kept consistent at 2.5 mm s−1, with a constant pressure of ~20 psi. The patterns were designed using Solidworks 3D design software (Dassault Systèmes SE). The 3D STL files were sliced using the slic3r engine from Repetier Host, which served as the main program for operating the extrusion bioprinter39.
Print fidelity test for the hydrogels (microbial ink)
Fidelity test namely filament collapse test was performed according to the previously published protocol31. A small structure with pillars featuring a series of spacing’s (2 L) was first 3D-printed. A single line (filament; green) of the bioink was extruded by the bioprinter from one side of the structure to the other end at a moderate nozzle moving speed of 5 mm s−1, suspending the bioink between the pillars (purple) and bridging the gaps. Photographs were taken from the side of the structure post-bioprinting to measure the structural integrity under gravitational force (Fg), in the form of angles (θ) of the overhung bioink fibers (Supplementary Fig. 14). Fidelity data fitting and theoretical modeling were conducted also in accordance with the previous report31. Data obtained from at least three independent samples were reported.
Photographs were taken from the side of the structure post-bioprinting to measure the structural integrity under gravitational force (Fg), in the form of angles (θ) of the overhung bioink fibers (Supplementary Fig. 14). Fidelity data fitting and theoretical modeling were conducted also in accordance with the previous report31. Data obtained from at least three independent samples were reported.
Preparation of functional microbial ink
The PQN4-Azu and PQN4-MazF cells were grown to OD 1 at 37 °C in 10 ml LB media, with 100 µg ml−1 carbenicillin and 50 µg ml−1 kanamycin, respectively. The 1 ml of microbial ink (CsgA-αγ) was evenly spread on the filter membrane and was incubated with 10 ml of bacterial culture (PQN4-Azu or PQN4-MazF) for 30 min to disperse the bacteria on the microbial ink and then vacuum filtered. In case of PQN4-BPA, the bacterial cells first were grown to OD 1 at 37 °C in 10 ml LB media with 100 µg ml−1 carbenicillin and 2% glucose (m v−1). Next, the cells were transferred to fresh LB media containing 100 µg ml−1 carbenicillin and the CsgA-BPA fibers were expressed by induction with 0.1 mM IPTG. After 24 h of expression, the engineered PQN4-BPA biofilm was incorporated to 1 ml of microbial ink (CsgA-αγ), as described above. The as-prepared microbial ink embedded with programmed bacterial cells/biofilms were utilized for 3D printing.
Next, the cells were transferred to fresh LB media containing 100 µg ml−1 carbenicillin and the CsgA-BPA fibers were expressed by induction with 0.1 mM IPTG. After 24 h of expression, the engineered PQN4-BPA biofilm was incorporated to 1 ml of microbial ink (CsgA-αγ), as described above. The as-prepared microbial ink embedded with programmed bacterial cells/biofilms were utilized for 3D printing.
Detection of secreted azurin
The 3D printed structures of microbial ink (CsgA-αγ) embedded with PQN4-Azu cells were immersed in LB media containing 100 µg ml−1 carbenicillin with or without 0.1 mM IPTG and incubated at 37 °C for 48 h. After 24 and 48 h of incubation, 0.5 ml of the culture media was collected for detection of azurin secreted by the microbial ink embedded with PQN4-Azu cells. The culture was concentrated on Amicon® Ultra-0.5 centrifugal filter (MiliporeSigma) to 20 µl. The protein present in concentrated supernatant were separated by NuPAGE 4–20% Tris-Glycine gels (ThermoFisher Scientific) and transferred to a nitrocellulose membrane using iBlot™ 2 Gel Transfer Device (ThermoFisher Scientific). The membrane was incubated with Goat Polyclonal Anti-Azurin Antibody (Origene CAT#: AB0048-100, Dilution 1:1000) at 4 °C overnight. After washing, the membranes were incubated with a secondary antibody—rabbit anti-goat IgG-HRP (abcam6741, Dilution 1:1000) for 1 h at room temperature. Chemiluminescence was detected using a FluorChem M system (ProteinSimple). Data obtained from at least three independent samples were reported.
The membrane was incubated with Goat Polyclonal Anti-Azurin Antibody (Origene CAT#: AB0048-100, Dilution 1:1000) at 4 °C overnight. After washing, the membranes were incubated with a secondary antibody—rabbit anti-goat IgG-HRP (abcam6741, Dilution 1:1000) for 1 h at room temperature. Chemiluminescence was detected using a FluorChem M system (ProteinSimple). Data obtained from at least three independent samples were reported.
BPA binding analysis
The 3D printed structures of microbial ink (CsgA-αγ) embedded with PQN4-BPA biofilm were immersed in water with or without 1 mM BPA and incubated for 24 h. After 12 and 24 h the water spiked with BPA was collected to test BPA binding by the microbial ink embedded with PQN4-BPA biofilm. The presence of BPA was detected using Agilent 6460 Triple Quad LC/MS and Agilent 1290 Infinity HPLC by Small Molecule Mass Spectrometry Facility at Harvard University, MA, USA. Data obtained from at least three independent samples were reported.
Cell growth under MazF
The 3D printed structures of microbial ink (CsgA-αγ) embedded with PQN4-MazF cells were immersed with LB media consisting of 100 µg ml−1 carbenicillin, with or without 0.1 mM IPTG at 37 °C for 6 h. The growth of PQN4-MazF cells in the microbial ink was monitored by collecting ~200 mg of the ink after 0, 2, 4, and 6 h. The collected microbial inks were transferred to the Eppendorf tubes and 1 ml of LB media was added to resuspend the ink, which was serially diluted, and plated on carbenicillin selective plates to obtain their CFU counts. Data obtained from at least three independent samples were reported.
Statistics and reproducibility
All experiments presented in this Article were repeated at least three times (n ≥ 3) on distinct samples, as clearly specified in the figure legends or the relevant Methods sections. In all cases, data are presented as the mean and standard deviation. GraphPad Prism 8 software was used for plotting and analyzing data. For micrographs and optical images, we present representative images.
For micrographs and optical images, we present representative images.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All relevant data supporting the findings of this study are available within the Article and its Supplementary Information or from the corresponding authors upon request. Source data are provided with this paper.
References
Murphy, S. V. & Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785 (2014).
Article CAS Google Scholar
Heinrich, M. A. et al. 3D Bioprinting: from benches to translational applications. Small 15, e1805510 (2019).
Article Google Scholar
Qian, F. et al. Direct writing of tunable living inks for bioprocess intensification. Nano Lett. 19, 5829–5835 (2019).
Article ADS CAS Google Scholar
Schaffner, M., Ruhs, P. A., Coulter, F., Kilcher, S. & Studart, A. R. 3D printing of bacteria into functional complex materials. Sci. Adv. 3, eaao6804 (2017).
Article Google Scholar
Gonzalez, L. M., Mukhitov, N. & Voigt, C. A. Resilient living materials built by printing bacterial spores. Nat. Chem. Biol. 16, 126–133 (2020).
Article CAS Google Scholar
Liu, X. et al. 3D printing of living responsive materials and devices. Adv. Mater. https://doi.org/10.1002/adma.201704821 (2018).
Lehner, B. A. E.
 , Schmieden, D. T. & Meyer, A. S. A straightforward approach for 3D bacterial printing. ACS Synth. Biol. 6, 1124–1130 (2017).
, Schmieden, D. T. & Meyer, A. S. A straightforward approach for 3D bacterial printing. ACS Synth. Biol. 6, 1124–1130 (2017).Article CAS Google Scholar
Huang, J. et al. Programmable and printable Bacillus subtilis biofilms as engineered living materials. Nat. Chem. Biol. 15, 34–41 (2019).
Article ADS CAS Google Scholar
Connell, J. L., Ritschdorff, E. T., Whiteley, M. & Shear, J. B. 3D printing of microscopic bacterial communities. Proc. Natl Acad. Sci. USA 110, 18380–18385 (2013).
Article ADS CAS Google Scholar
Smith, R. S. H. et al. Hybrid living materials: digital design and fabrication of 3D multimaterial structures with programmable biohybrid surfaces. Adv.
 Funct. Mater. 30, 1907401 (2020).
Funct. Mater. 30, 1907401 (2020).Article CAS Google Scholar
Schwab, A. et al. Printability and shape fidelity of bioinks in 3D bioprinting. Chem. Rev. 120, 11028–11055 (2020).
Article CAS Google Scholar
Joshi, S., Cook, E. & Mannoor, M. S. Bacterial nanobionics via 3D printing. Nano Lett. 18, 7448–7456 (2018).
Article ADS CAS Google Scholar
Saha, A. et al. Additive manufacturing of catalytically active living materials. ACS Appl Mater. Interfaces 10, 13373–13380 (2018).
Article CAS Google Scholar
Nguyen, P. Q., Courchesne, N. D., Duraj-Thatte, A., Praveschotinunt, P. & Joshi, N.
 S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, e1704847 (2018).
S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, e1704847 (2018).Article Google Scholar
Tang, T.-C. et al. Materials design by synthetic biology. Nat. Rev. Mater. https://doi.org/10.1038/s41578-020-00265-w (2020).
Gilbert, C. & Ellis, T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8, 1–15 (2019).
Article CAS Google Scholar
Chen, A. Y., Zhong, C. & Lu, T. K. Engineering living functional materials. Acs Synth. Biol. 4, 8–11 (2015).
Article Google Scholar
Duraj-Thatte, A.
 M. et al. Genetically programmable self-regenerating bacterial hydrogels. Adv. Mater. 31, e1901826 (2019).
M. et al. Genetically programmable self-regenerating bacterial hydrogels. Adv. Mater. 31, e1901826 (2019).Article Google Scholar
Manjula-Basavanna, A., Duraj-Thatte, A. M. & Joshi, N. S. Robust self-regeneratable stiff living materials fabricated from microbial cells. Adv. Funct. Mater. n/a, 2010784, https://doi.org/10.1002/adfm.202010784 (2021).
Duraj-Thatte, A. M. et al. Water-processable, biodegradable and coatable aquaplastic from engineered biofilms. Nat Chem. Biol. https://doi.org/10.1038/s41589-021-00773-y (2021).
Praveschotinunt, P. et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580 (2019).
Article ADS CAS Google Scholar
Gilbert, C. et al. Living materials with programmable functionalities grown from engineered microbial co-cultures. Nat. Mater. https://doi.org/10.1038/s41563-020-00857-5 (2021).
Article PubMed Google Scholar
Tay, P. K. R., Manjula-Basavanna, A. & Joshi, N. S. Repurposing bacterial extracellular matrix for selective and differential abstraction of rare earth elements. Green. Chem. 20, 3512–3520 (2018).
Article CAS Google Scholar
Birnbaum, D. P., Manjula-Basavanna, A., Kan, A., Tardy, B. L. & Joshi, N. S. Hybrid living capsules autonomously produced by engineered bacteria. Adv. Sci. https://doi.org/10.1002/advs.202004699 (2021).
Nguyen, P. Q., Botyanszki, Z., Tay, P. K. & Joshi, N. S. Programmable biofilm-based materials from engineered curli nanofibres.
 Nat. Commun. 5, 4945 (2014).
Nat. Commun. 5, 4945 (2014).Article ADS CAS Google Scholar
Chen, A. Y. et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523 (2014).
Article ADS CAS Google Scholar
Zhong, C. et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 9, 858–866 (2014).
Article ADS CAS Google Scholar
Moser, F., Tham, E., Gonzalez, L. M., Lu, T. K. & Voigt, C. A. Light-controlled, high-resolution patterning of living engineered bacteria onto textiles, ceramics, and plastic. Adv. Funct. Mater. https://doi.org/10.1002/adfm.201901788 (2019).
Pratt, K.
 P., Cote, H. C., Chung, D. W., Stenkamp, R. E. & Davie, E. W. The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro. Proc. Natl Acad. Sci. USA 94, 7176–7181 (1997).
P., Cote, H. C., Chung, D. W., Stenkamp, R. E. & Davie, E. W. The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro. Proc. Natl Acad. Sci. USA 94, 7176–7181 (1997).Article ADS CAS Google Scholar
Dorval Courchesne, N. M., Duraj-Thatte, A., Tay, P. K. R., Nguyen, P. Q. & Joshi, N. S. Scalable production of genetically engineered nanofibrous macroscopic materials via filtration. ACS Biomater. Sci. Eng. 3, 733–741 (2017).
Article CAS Google Scholar
Ribeiro, A. et al. Assessing bioink shape fidelity to aid material development in 3D bioprinting. Biofabrication 10, 014102 (2017).
Article ADS CAS Google Scholar
Highley, C. B., Rodell, C. B. & Burdick, J. A. Direct 3D printing of shear-thinning hydrogels into self-healing hydrogels. Adv. Mater. 27, 5075–5079 (2015).
Article CAS Google Scholar
Liu, W. et al. Extrusion bioprinting of shear-thinning gelatin methacryloyl bioinks. Adv. Healthc. Mater. https://doi.org/10.1002/adhm.201601451 (2017).
Yamada, T. et al. Apoptosis or growth arrest: Modulation of tumor suppressor p53’s specificity by bacterial redox protein azurin. Proc. Natl Acad. Sci. USA 101, 4770–4775 (2004).
Article ADS CAS Google Scholar
Maruthamuthu, M. K. et al. Development of bisphenol A-removing recombinant Escherichia coli by monomeric and dimeric surface display of bisphenol A-binding peptide. Bioprocess Biosyst.
 Eng. 41, 479–487 (2018).
Eng. 41, 479–487 (2018).Article CAS Google Scholar
Engelberg-Kulka, H., Hazan, R. & Amitai, S. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 118, 4327–4332 (2005).
Article CAS Google Scholar
Rothschild, L. J. Synthetic biology meets bioprinting: enabling technologies for humans on Mars (and Earth). Biochem. Soc. Trans. 44, 1158–1164 (2016).
Article CAS Google Scholar
Kan, A., Birnbaum, D. P., Praveschotinunt, P. & Joshi, N. S. Congo red fluorescence for rapid in situ characterization of synthetic curli systems. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.00434-19 (2019).
Ying, G.
 L. et al. Aqueous two-phase emulsion bioink-enabled 3D bioprinting of porous hydrogels. Adv. Mater. 30, e1805460 (2018).
L. et al. Aqueous two-phase emulsion bioink-enabled 3D bioprinting of porous hydrogels. Adv. Mater. 30, e1805460 (2018).Article Google Scholar
Download references
Acknowledgements
Work was performed in part at the Center for Nanoscale Systems at Harvard. Work in the N.S.J. laboratory is supported by the National Institutes of Health (1R01DK110770), the National Science Foundation (DMR 2004875), and the Wyss Institute for Biologically Inspired Engineering at Harvard University. Work in the D.A.W. laboratory is supported by the Harvard University Materials Research Science and Engineering Center (NSF Grants DMR-1420570 and DMR-2011754). Work in the Y.S.Z. laboratory was supported by the Lush Prize and the Brigham Research Institute. Parts of the schematics were adapted from BioRender.com.
Author information
Author notes
Anna M. Duraj-Thatte
Present address: Department of Biological Systems Engineering, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA
These authors contributed equally: Anna M.
 Duraj-Thatte, Avinash Manjula-Basavanna.
Duraj-Thatte, Avinash Manjula-Basavanna.
Authors and Affiliations
John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA, USA
Anna M. Duraj-Thatte, Jing Xia, David A. Weitz & Neel S. Joshi
Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, USA
Anna M. Duraj-Thatte, Avinash Manjula-Basavanna, Jarod Rutledge, Arjirios Sourlis, Anton Kan & Neel S. Joshi
Department of Chemistry and Chemical Biology, Northeastern University, Boston, MA, USA
Anna M. Duraj-Thatte, Avinash Manjula-Basavanna & Neel S. Joshi
Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA, USA
Shabir Hassan, Andrés G. Rubio, Ami Lesha, Michael Zenkl & Yu Shrike Zhang
Authors
- Anna M. Duraj-Thatte
View author publications
You can also search for this author in PubMed Google Scholar
- Avinash Manjula-Basavanna
View author publications
You can also search for this author in PubMed Google Scholar
- Jarod Rutledge
View author publications
You can also search for this author in PubMed Google Scholar
- Jing Xia
View author publications
You can also search for this author in PubMed Google Scholar
- Shabir Hassan
View author publications
You can also search for this author in PubMed Google Scholar
- Arjirios Sourlis
View author publications
You can also search for this author in PubMed Google Scholar
- Andrés G.
 Rubio
RubioView author publications
You can also search for this author in PubMed Google Scholar
- Ami Lesha
View author publications
You can also search for this author in PubMed Google Scholar
- Michael Zenkl
View author publications
You can also search for this author in PubMed Google Scholar
- Anton Kan
View author publications
You can also search for this author in PubMed Google Scholar
- David A. Weitz
View author publications
You can also search for this author in PubMed Google Scholar
- Yu Shrike Zhang
View author publications
You can also search for this author in PubMed Google Scholar
- Neel S. Joshi
View author publications
You can also search for this author in PubMed Google Scholar
Contributions
A. M.D.-T. and A.M.-B. conceived the project. A.M.D.-T. and J.R. cloned CsgA-α and CsgA-γ. A.M.D.-T., A.M.-B., A.S., and J.R. produced hydrogel-based microbial inks. A.M.-B. did FESEM and TEM imaging. J.X. tested the rheological properties of the microbial inks. S.H., A.G.R., A.L., and M.Z. tested printing performance and printed 3D structures. D.A.W. supervised J.X. Y.S.Z. supervised S.H., A.G.R., A.L., and M.Z. A.M.D.-T. cloned PQN4-Azu and PQN4-BPA. A.K. cloned PQN4-MazF. A.M.D.-T. and A.M.-B tested and analyzed living materials produced from PQN4-Azu, PQN4-BPA, and PQN4-MazF. A.M.-B., A.M.D.-T., and N.S.J. wrote and/or edited the manuscript. All authors discussed and commented on the manuscript.
M.D.-T. and A.M.-B. conceived the project. A.M.D.-T. and J.R. cloned CsgA-α and CsgA-γ. A.M.D.-T., A.M.-B., A.S., and J.R. produced hydrogel-based microbial inks. A.M.-B. did FESEM and TEM imaging. J.X. tested the rheological properties of the microbial inks. S.H., A.G.R., A.L., and M.Z. tested printing performance and printed 3D structures. D.A.W. supervised J.X. Y.S.Z. supervised S.H., A.G.R., A.L., and M.Z. A.M.D.-T. cloned PQN4-Azu and PQN4-BPA. A.K. cloned PQN4-MazF. A.M.D.-T. and A.M.-B tested and analyzed living materials produced from PQN4-Azu, PQN4-BPA, and PQN4-MazF. A.M.-B., A.M.D.-T., and N.S.J. wrote and/or edited the manuscript. All authors discussed and commented on the manuscript.
Corresponding authors
Correspondence to Anna M. Duraj-Thatte, Avinash Manjula-Basavanna or Neel S. Joshi.
Ethics declarations
Competing interests
A.M.D.-T., A.M.-B., A.S., and N.S.J. are inventors on a U.S. Patent Application No. 17/254,019 submitted by Harvard University. The other authors declare no competing interests.
17/254,019 submitted by Harvard University. The other authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Video 1
Description of Additional Supplementary Files
Reporting Summary
Source data
Source Data
Peer Review File
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
3D printing of bacteria into functional complex materials
1. Prosser J. I., Bohannan B. J. M., Curtis T. P., Ellis R. J., Firestone M. K., Freckleton R. P., Green J. L., Green L. E., Killham K., Lennon J. J., Mark Osborn A., Solan M., van der Gast C. J., Young J. P. W., The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392 (2007). [PubMed] [Google Scholar]
2. Hall-Stoodley L., Costerton J. W., Stoodley P. , Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [PubMed] [Google Scholar]
, Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [PubMed] [Google Scholar]
3. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M., Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995). [PubMed] [Google Scholar]
4. Flemming H.-C., Wingender J., The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [PubMed] [Google Scholar]
5. Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S., Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). [PubMed] [Google Scholar]
6. Branda S. S., Vik Å., Friedman L., Kolter R., Biofilms: The matrix revisited. Trends Microbiol. 13, 20–26 (2005). [PubMed] [Google Scholar]
7. Demain A. L., Microbial biotechnology. Trends Biotechnol. 18, 26–31 (2000). [PubMed] [Google Scholar]
8. Douglas S., Beveridge T. J., Mineral formation by bacteria in natural microbial communities. FEMS Microbiol. Ecol. 26, 79–88 (1998). [Google Scholar]
FEMS Microbiol. Ecol. 26, 79–88 (1998). [Google Scholar]
9. Bazylinski D. A., Frankel R. B., Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230 (2004). [PubMed] [Google Scholar]
10. Rehm B. H. A., Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592 (2010). [PubMed] [Google Scholar]
11. Klaus-Joerger T., Joerger R., Olsson E., Granqvist C.-G., Bacteria as workers in the living factory: Metal-accumulating bacteria and their potential for materials science. Trends Biotechnol. 19, 15–20 (2001). [PubMed] [Google Scholar]
12. Bhuwal A. K., Singh G., Aggarwal N. K., Goyal V., Yadav A., Isolation and screening of polyhydroxyalkanoates producing bacteria from pulp, paper, and cardboard industry wastes. Int. J. Biomater. 2013, 752821 (2013). [PMC free article] [PubMed] [Google Scholar]
13. Bottan S., Robotti F., Jayathissa P., Hegglin A., Bahamonde N., Heredia-Guerrero J. A., Bayer I. S., Scarpellini A. , Merker H., Lindenblatt N., Poulikakos D., Ferrari A., Surface-structured bacterial cellulose with guided assembly-based biolithography (GAB). ACS Nano 9, 206–219 (2015). [PubMed] [Google Scholar]
, Merker H., Lindenblatt N., Poulikakos D., Ferrari A., Surface-structured bacterial cellulose with guided assembly-based biolithography (GAB). ACS Nano 9, 206–219 (2015). [PubMed] [Google Scholar]
14. Cassidy M. B., Lee H., Trevors J. T., Environmental applications of immobilized microbial cells: A review. J. Ind. Microbiol. 16, 79–101 (1996). [Google Scholar]
15. Rühs P. A., Böni L., Fuller G. G., Inglis R. F., Fischer P., In-situ quantification of the interfacial rheological response of bacterial biofilms to environmental stimuli. PLOS ONE 8, e78524 (2013). [PMC free article] [PubMed] [Google Scholar]
16. Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R., Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013). [PMC free article] [PubMed] [Google Scholar]
17. Brown R. M. Jr, Willison J. H., Richardson C. L., Cellulose biosynthesis in Acetobacter xylinum: Visualization of the site of synthesis and direct measurement of the in vivo process. Proc. Natl. Acad. Sci. U.S.A. 73, 4565–4569 (1976). [PMC free article] [PubMed] [Google Scholar]
Proc. Natl. Acad. Sci. U.S.A. 73, 4565–4569 (1976). [PMC free article] [PubMed] [Google Scholar]
18. Bäckdahl H., Helenius G., Bodin A., Nannmark U., Johansson B. R., Risberg B., Gatenholm P., Mechanical properties of bacterial cellulose and interactions with smooth muscle cells. Biomaterials 27, 2141–2149 (2006). [PubMed] [Google Scholar]
19. Chin N. L., Man H. C., Talib R. A., 2nd international conference on agricultural and food engineering (CAFEi 2014): New trends forward. Agric. Agric. Sci. Procedia 2, 1–414 (2014). [Google Scholar]
20. Shi Z., Zhang Y., Phillips G. O., Yang G., Utilization of bacterial cellulose in food. Food Hydrocoll. 35, 539–545 (2014). [Google Scholar]
21. Sulaeva I., Henniges U., Rosenau T., Potthast A., Bacterial cellulose as a material for wound treatment: Properties and modifications. A review. Biotechnol. Adv. 33, 1547–1571 (2015). [PubMed] [Google Scholar]
22. Czaja W. K., Young D. J., Kawecki M., Brown R. M. Jr, The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 8, 1–12 (2007). [PubMed] [Google Scholar]
M. Jr, The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 8, 1–12 (2007). [PubMed] [Google Scholar]
23. Nimeskern L., Ávila H. M., Sundberg J., Gatenholm P., Müller R., Stok K. S., Mechanical evaluation of bacterial nanocellulose as an implant material for ear cartilage replacement. J. Mech. Behav. Biomed. Mater. 22, 12–21 (2013). [PubMed] [Google Scholar]
24. Kowalska-Ludwicka K., Cala J., Grobelski B., Sygut D., Jesionek-Kupnicka D., Kolodziejczyk M., Bielecki S., Pasieka Z., Modified bacterial cellulose tubes for regeneration of damaged peripheral nerves. Arch. Med. Sci. 9, 527–534 (2013). [PMC free article] [PubMed] [Google Scholar]
25. Logan B. E., Hamelers B., Rozendal R., Schröder U., Keller J., Freguia S., Aelterman P., Verstraete W., Rabaey K., Microbial fuel cells: Methodology and technology. Environ. Sci. Technol. 40, 5181–5192 (2006). [PubMed] [Google Scholar]
26. Su L., Jia W., Hou C., Lei Y., Microbial biosensors: A review. Biosens. Bioelectron. 26, 1788–1799 (2011). [PubMed] [Google Scholar]
Biosens. Bioelectron. 26, 1788–1799 (2011). [PubMed] [Google Scholar]
27. Connell J. L., Kim J., Shear J. B., Bard A. J., Whiteley M., Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. U.S.A. 111, 18255–18260 (2014). [PMC free article] [PubMed] [Google Scholar]
28. Connell J. L., Ritschdorff E. T., Whiteley M., Shear J. B., 3D printing of microscopic bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 110, 18380–18385 (2013). [PMC free article] [PubMed] [Google Scholar]
29. Bodelón G., Montes-García V., López-Puente V., Hill E. H., Hamon C., Sanz-Ortiz M. N., Rodal-Cedeira S., Costas C., Celiksoy S., Pérez-Juste I., Scarabelli L., La Porta A., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L. M., Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat. Mater. 15, 1203–1211 (2016). [PMC free article] [PubMed] [Google Scholar]
30. Lehner B. A. E., Schmieden D. T., Meyer A. S., A straightforward approach for 3D bacterial printing. ACS Synth. Biol. 6, 1124–1130 (2017). [PMC free article] [PubMed] [Google Scholar]
Lehner B. A. E., Schmieden D. T., Meyer A. S., A straightforward approach for 3D bacterial printing. ACS Synth. Biol. 6, 1124–1130 (2017). [PMC free article] [PubMed] [Google Scholar]
31. Bader C., Patrick W. G., Kolb D., Hays S. G., Keating S., Sharma S., Dikovsky D., Belocon B., Weaver J. C., Silver P. A., Oxman N., Grown, printed, and biologically augmented: An additively manufactured microfluidic wearable, functionally templated for synthetic microbes. 3D Print. Addit. Manuf. 3, 79–89 (2016). [Google Scholar]
32. Kokkinis D., Schaffner M., Studart A. R., Multimaterial magnetically assisted 3D printing of composite materials. Nat. Commun. 6, 8643 (2015). [PMC free article] [PubMed] [Google Scholar]
33. P. Gatenholm, H. Backdahl, T. J. Tzavaras, R. V Davalos, M. B. Sano, “Three-dimensional bioprinting of biosynthetic cellulose (BC) implants and scaffolds for tissue engineering,” U.S. Patent 8,691,974 (2014).
34. Fu Q., Saiz E., Tomsia A. P., Direct ink writing of highly porous and strong glass scaffolds for load-bearing bone defects repair and regeneration. Acta Biomater. 7, 3547–3554 (2011). [PMC free article] [PubMed] [Google Scholar]
Acta Biomater. 7, 3547–3554 (2011). [PMC free article] [PubMed] [Google Scholar]
35. Lewis J. A., Direct ink writing of 3D functional materials. Adv. Funct. Mater. 16, 2193–2204 (2006). [Google Scholar]
36. Smay J. E., Cesarano J. III, Lewis J. A., Colloidal inks for directed assembly of 3-D periodic structures. Langmuir 18, 5429–5437 (2002). [Google Scholar]
37. Minas C., Carnelli D., Tervoort E., Studart A. R., 3D printing of emulsions and foams into hierarchical porous ceramics. Adv. Mater. 28, 9993–9999 (2016). [PubMed] [Google Scholar]
38. Rose S., Prevoteau A., Elzière P., Hourdet D., Marcellan A., Leibler L., Nanoparticle solutions as adhesives for gels and biological tissues. Nature 505, 382–385 (2014). [PubMed] [Google Scholar]
39. Jungst T., Smolan W., Schacht K., Scheibel T., Groll J., Strategies and molecular design criteria for 3D printable hydrogels. Chem. Rev. 116, 1496–1539 (2016). [PubMed] [Google Scholar]
40. Lee Y., Chung H. J., Yeo S., Ahn C.-H., Lee H., Messersmith P. B., Park T. G., Thermo-sensitive, injectable, and tissue adhesive sol–gel transition hyaluronic acid/pluronic composite hydrogels prepared from bio-inspired catechol-thiol reaction. Soft Matter 6, 977–983 (2010). [Google Scholar]
J., Yeo S., Ahn C.-H., Lee H., Messersmith P. B., Park T. G., Thermo-sensitive, injectable, and tissue adhesive sol–gel transition hyaluronic acid/pluronic composite hydrogels prepared from bio-inspired catechol-thiol reaction. Soft Matter 6, 977–983 (2010). [Google Scholar]
41. Hyun K., Wilhelm M., Klein C. O., Cho K. S., Nam J. G., Ahn K. H., Lee S. J., Ewoldt R. H., McKinley G. H., A review of nonlinear oscillatory shear tests: Analysis and application of large amplitude oscillatory shear (LAOS). Prog. Polym. Sci. 36, 1697–1753 (2011). [Google Scholar]
42. Allsop P. J., Chisti Y., Moo-Young M., Sullivan G. R., Dynamics of phenol degradation by Pseudomonas putida. Biotechnol. Bioeng. 41, 572–580 (1993). [PubMed] [Google Scholar]
43. Yang R. D., Humphrey A. E., Dynamic and steady state studies of phenol biodegradation in pure and mixed cultures. Biotechnol. Bioeng. 17, 1211–1235 (1975). [PubMed] [Google Scholar]
44. Hasan S. A., Jabeen S., Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnol. Biotechnol. Equip. 29, 45–53 (2015). [PMC free article] [PubMed] [Google Scholar]
Biotechnol. Biotechnol. Equip. 29, 45–53 (2015). [PMC free article] [PubMed] [Google Scholar]
45. Das A. A. K., Bovill J., Ayesh M., Stoyanov S. D., Paunov V. N., Fabrication of living soft matter by symbiotic growth of unicellular microorganisms. J. Mater. Chem. B 4, 3685–3694 (2016). [Google Scholar]
46. Tittsler R. P., Sandholzer L. A., The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31, 575–580 (1936). [PMC free article] [PubMed] [Google Scholar]
47. Leach J. B., Schmidt C. E., Characterization of protein release from photocrosslinkable hyaluronic acid-polyethylene glycol hydrogel tissue engineering scaffolds. Biomaterials 26, 125–135 (2005). [PubMed] [Google Scholar]
48. Bertucco A., Spilimbergo S., Treating micro-organisms with high pressure. Ind. Chem. Libr. 9, 626–640 (2001). [Google Scholar]
3D printing bacteria and electronics of the future. 3D technologies
"Wearable technology", which is introduced into the human body, is no longer considered a fantasy. And now, thanks to the use of the natural reaction of bacteria to a specific stimulus, wearable equipment can work without electricity.
And now, thanks to the use of the natural reaction of bacteria to a specific stimulus, wearable equipment can work without electricity.
The Massachusetts Institute of Technology is actively engaged in all kinds of innovations. The portfolio of materials developed by MIT has been replenished with “smart living ink”. More recently, research scientists at MIT have developed bio-inks based on living cells that are used for 3D printing. This is a truly "smart ink" that changes ideas about medicine, electronics and the use of 3D technologies by 180 degrees.
Bacterial 3D printer ink was developed by the makers of robotic soft gloves used for fishing. Research is associated with the names of such scientists as Professor Xuan Zhao and Dr. Timothy Lu. They work in a laboratory that develops soft active materials. These scientists conducted research in the programming of genetically engineered bacteria. To confirm their concept, scientists used bioink with bacteria that were genetically modified so that when interacting with a certain kind of chemical signal, they began to glow.
The ink is made from special ingredients that are selected to form the ideal environment for living cells. The hydrogel base is mixed with a photosensitizer that guarantees setting, bacterial cell pellets, nutrients and deionized water.
The first attempts did not bring the desired result, since at first the scientists used mammalian cells that could not withstand the 3D printing process and died. The fact is, as the scientists explained, that these cells are essentially two-layer lipid bubbles, that is, they are too weak and prone to easy destruction. Others were selected, which turned out to be bacterial cells - they met all the requirements. The walls of bacteria are very strong and can withstand significant pressure that occurs during 3D printing. In addition, they live well in an aquatic hydrogel environment.
In the course of research, bacterial cells were genetically modified in such a way that, in response to a certain chemical signal, the cells began to produce green fluorescent proteins.
To begin with, the scientists designed geometric structures in SOLIDWORKS and CADfusion in the form of a square, a pyramid, a pyramid with voids, and a hemisphere. Then they processed them with a slicer in the program and transferred them to a 3D printer, which printed them with living bacteria-based ink. The result is small figures, but they are perfectly visible to the naked eye. The dimensions of the figures were 3 cm, and the print accuracy was 30 μm.
3D printing was done using Aerotech, a three-axis robotic application technology. Printing was carried out without creating certain temperature conditions. Hydrogel-based ink from the UV-impervious body of the syringe was fed into the working area using nozzles, the diameter of which was 30-250 μm. A layer of bio-ink was applied to the cross-array to ensure cell viability and sensitivity to the living network. Subsequently, the hydrogel was subjected to UV irradiation, due to which a cross chemical compound was formed. At the end of the process, the bioink was placed in a high humidity environment.
At the end of the process, the bioink was placed in a high humidity environment.
To demonstrate how signal chemicals can activate the production of fluorescent proteins on different surfaces, a pyramid was printed using different bacterial strains.
In the course of the experiments, the structures were exposed to a chemical signal, which provoked the coloring of certain parts of the pyramid: the top and bottom of the pyramid became green, while the lateral parts did not change their initial red color.
Scientists went further and created a "living tattoo". Based on three types of hydrogel inks based on different strains of bacteria, tattoos were printed that reflect certain chemical signals present on human skin.
The tattoo was made in the form of a tree pattern applied to a thin elastomer. After printing, the tattoo was pasted onto the skin. Individual branches of the tattoo glowed, depending on the presence of a chemical irritant on the skin to which the bacteria reacted.
Living ink can also be used in the medical field. In particular, such inks, or rather the bacterial cells in them, can be used as workers in a factory. Genetic engineering makes it possible for cells to produce drugs in a 3D platform under the influence of a certain reagent, and they can be used not only as a subcutaneous device. As long as it is possible to maintain the viability of bacteria, this method can be used to print implants or food.
3D printing with living hydrogel ink can also be used in the field of computers. In particular, it is possible to replace the electronic interaction of microcomponents in chips with the programmed interaction of cells and chemicals. As a result, you can solve logical problems and perform Boolean operations.
In the classical scheme, there are elements at the input, which will correspond to the value 0 or 1. The ink lines can be arranged in such a way that the presence of active components or their absence will correspond to these values. The same can be done on the output, that is, the presence of the corresponding proteins will correspond to 1 or 0. This means that it is possible to program the material.
The same can be done on the output, that is, the presence of the corresponding proteins will correspond to 1 or 0. This means that it is possible to program the material.
In the general structure of the material, each single cell can perform a computational operation. But if you combine these cells in space into a certain three-dimensional structure, then interaction in different areas can give rise to information structures that can solve very complex problems. This means that three-dimensional chemical detectors and logic gates can be created using living ink.
Will 3D printed bacteria be the electronics of the future?
"Wearable technology" - the so-called "smart" electronic devices that can be worn on oneself or implanted in the body. Offering enticing prospects for innovative technology developers in sports, healthcare, fashion and entertainment, 3D printing opens up new possibilities for wearable technology such as second e-skin and smart fabrics.
In a recent study, a group of scientists from the Massachusetts Institute of Technology (MIT) has developed a "living" biological ink for a 3D printer, which is not only "smart", but also has the potential to completely change our understanding of this technology. Using the natural reactions of bacteria, devices made with such inks are the basic building blocks of wearable technology that does not use electricity.
This 3D printed bioink comes from the same MIT group that recently came up with soft robotic fishing gloves. The new ink expands the vast portfolio of smart materials being developed at MIT.
3D printing and programming of genetically engineered bacteria
This materials research at MIT was led by Prof. Xuanhe Zhao and Dr. Timothy Lu of the Soft Active Materials Laboratory. As a proof of concept, a team of scientists demonstrated how bacteria in 3D printer bioink can be genetically modified to glow when it receives a specific chemical signal.
First, the ink is prepared, the ingredients of which make it an ideal microenvironment for living matter. To the hydrogel base, the scientists added a photosensitizer that provides the setting of the material, as well as bacterial cell granules, nutrients and deionized water.
When preparing the ink, it was very important to choose the right bacteria, as previous attempts using other cells had failed. “It turned out that these cells died during the printing process, because mammalian cells are basically two-layered lipid bubbles,” explains Hyunwoo Yuk, one of the co-authors of the study. “They are too weak and break easily.”
Introduction of digital bacteria. Photo MIT Bacterial cells were chosen because the walls protecting them are able to withstand the pressure of the 3D printing process and live in the aquatic environment of hydrogels. The bacteria have been genetically modified to allow them to produce green fluorescent proteins (GFPs), either normally or when activated by a specific chemical signal.
3D printing of complex living structures with living ink
As a first example, scientists 3D printed complex structures visible to the naked eye (3 cm in size) with microscopic precision (30 microns).
This included a square, a solid pyramid, a pyramid with voids, and a dome. These complex designs were designed using SOLIDWORKS and CADfusion software, then cut into layers and printed at room temperature using Aerotech's 3-axis robotic application technology.
Hydrogel ink was dispensed from a 5 ml UV-impermeable syringe barrel through nozzles ranging in diameter from 30 to 250 µm.
Initially, the ink was applied by a 3d printer on a cross matrix. This design was necessary to ensure the viability and sensitivity of cells in a living network. Chemical crosslinking was then created by exposing the 3D printed hydrogel to UV light. Thereafter, the cross-linked hydrogel ink was placed in humid conditions.
To demonstrate how different signaling chemicals can activate GFP production in different areas of the mold, the scientists 3D printed a pyramid using two different strains of bacteria.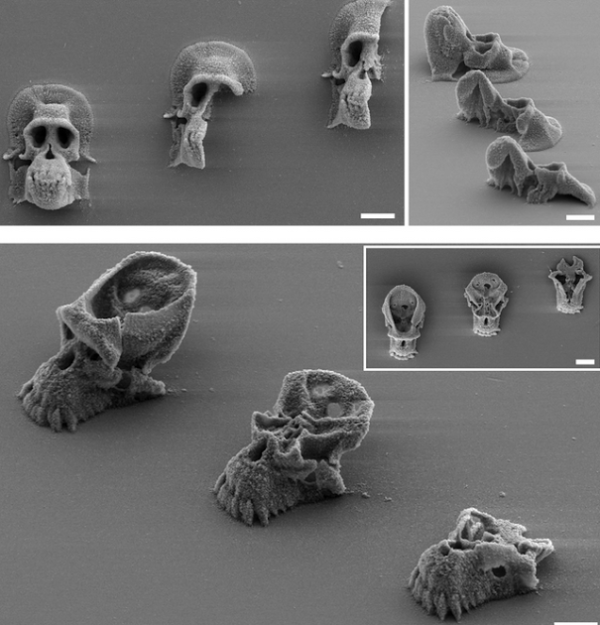
When exposed to a specific chemical signal, the top and base of the pyramid turned fluorescent green, while the sides remained red.
Complex objects 3d printed using living hydrogel ink. Photograph by VCH-Verlag.3d printing of living tattoos
The second example was the use of three different hydrogel inks, using different strains of bacteria. Here, 3D printing has been used to create a "living tattoo" that reflects the presence of certain chemical signals on the skin.
The tattoo was printed with cells of living bacteria, and was a tree-like pattern on a thin layer of elastomer. Then it was glued to human skin.
Three branches of a tree were 3D printed with different strains of live bacteria that respond to different chemical signaling compounds. Within hours, tree branches began to glow when the bacteria reacted to the appropriate chemical irritant on the skin.
Diagram showing a 3D printed tattoo with different skin stretch (b). Illumination with GFP as a result of a reaction in different areas of the skin (c). Photograph by VCH-Verlag.
Photograph by VCH-Verlag. Possible computer applications for 3d printing of hydrogel inks
The third possibility of using hydrogel inks in 3d printing is computational operations. Instead of the interaction of electronic components in a computer chip, it is possible to program the interaction of cells and chemical compounds, creating logic gates to perform Boolean operations.
The ink lines are arranged in such a way that instead of an electronic input, a value of 0 or 1 corresponds to the absence or presence of active components. Similarly, the absence or presence of produced GFPs corresponds to a value of 0 or 1 on the output. In this way, you can program the material.
“Each cell in the structure performs a simple computational operation,” Lu explains. “However, when spatially combined into three-dimensional structures, the interaction between different cell types and chemical compounds in different regions can generate informative circuits and provide complex logical calculations.