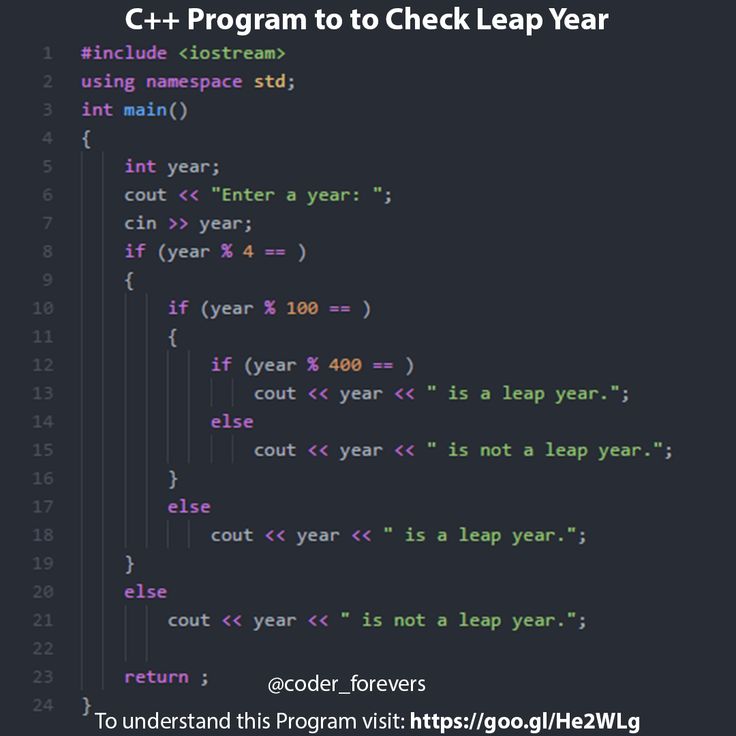
3D printing dna
New Research Could Lead to DNA 3D Printer - 3DPrint.com
Technology is capable of amazing things, but it doesn’t mean those things are easy. It’s incredible that scientists can produce DNA in a lab, but the process is difficult, lengthy and requires toxic chemicals. Imagine, however, if they could simply print it, the way that you would 3D print anything else. That could be the future, after scientists at UC Berkeley and Lawrence Berkeley National Laboratory developed a new way to synthesize DNA. The method could lead to DNA printers, similar to ordinary 3D printers, that could produce DNA strands that are more accurate and 10 times longer than the strands produced with today’s methods – more quickly and easily, and without the use of toxic chemicals.
“If you’re a mechanical engineer, it’s really nice to have a 3D printer in your shop that can print out a part overnight so you can test it the next morning,” said UC Berkeley graduate student Dan Arlow. “If you’re a researcher or bioengineer and you have an instrument that streamlines DNA synthesis, a ‘DNA printer,’ you can test your ideas faster and try out more new ideas.
I think it will lead to a lot of innovation.”
The research was led by Arlow and PhD student Sebastian Palluk, a doctoral student at the Technische Universität Darmstadt in Germany and a visiting student at Berkeley Lab. It is published in a paper entitled “De novo DNA synthesis using polymerase-nucleotide conjugates,” which you can access here. The research was conducted at the Department of Energy’s Joint BioEnergy Institute (JBEI).
“I personally think Dan and Sebastian’s new method could revolutionize how we make DNA,” said Jay Keasling, a UC Berkeley professor of chemical and biomolecular engineering, senior faculty scientist at Berkeley Lab and Chief Executive Officer of JBEI.
Keasling and JBEI scientists specialize specialize in adding genes to microbes, typically yeast and bacteria, to sustainably produce useful products. Palluk came from Germany specifically to work with Arlow in Keasling’s lab.
L to R: Dan Arlow, Sebastian Palluk and Jay Keasling
“We believe that increased access to DNA constructs will speed up the development of new cures for diseases and simplify the production of new medicines,” Palluk said.

The synthesis of DNA is a growing business; companies are ordering custom-made genes so that they can produce chemicals, biologic drugs or industrial enzymes. Researchers purchase synthetic genes to engineer plants and animals or try out new CRISPR-based disease therapies. Some scientists have even researched storing information in DNA, but that would require much larger quantities of DNA than are currently synthesized. All of these applications require that synthesis produce the desired sequence of nucleotides or bases, the building blocks of DNA, in each of millions or billions of copies of DNA molecules.
Current DNA synthesis is limited to producing oligonucleotides about 200 bases long, because errors in the process lead to a low yield of correct sequences as the length increases. To assemble even a small gene, scientists have to stitch together segments of about 200 bases long. The turnaround time for a small gene of 1,500 bases long can be two weeks at a cost of $300, limiting the experiments that scientists can do. Synthetic biologists like Arlow, Palluk and Keasling often insert a dozen different genes at once into a microbe to get it to produce a chemical, and each gene presents its own synthesis problems.
Synthetic biologists like Arlow, Palluk and Keasling often insert a dozen different genes at once into a microbe to get it to produce a chemical, and each gene presents its own synthesis problems.
“As a student in Germany, I was part of an international synthetic biology competition, iGEM, where we tried to get E. coli bacteria to degrade plastic waste,” said Palluk. “But I soon realized that most of the research time was spent just getting all the DNA together, not doing the experiments to see if the engineered cells could break down the plastic. This really motivated me to look into the DNA synthesis process.”
The technology developed by Palluk, Arlow, Keasling and their team relies on a DNA-synthesizing enzyme found in cells of the immune system that has the natural ability to add nucleotides to an existing DNA molecule in water, where DNA is most stable. The technology results in increased precision, allowing synthesis of DNA strands several thousand bases long – a medium-sized gene.
“We have come up with a novel way to synthesize DNA that harnesses the machinery that nature itself uses to make DNA,” Palluk said. “This approach is promising because enzymes have evolved for millions of years to perform this exact chemistry.”
Cells create DNA by copying it with the help of several different polymerase enzymes that build on DNA already in the cell. But in the 1960s, scientists discovered a polymerase that doesn’t rely on an existing DNA template but instead randomly adds nucleotides to genes that make antibodies for the immune system. The enzyme, called terminal deoxynucleotidyl transferase (TdT), creates random variation in these genes, resulting in antibody proteins that are better able to attack new types of invaders.
TdT is fast and does not have side-reactions that could affect the resulting molecule. Scientists over the years have tried to use the enzyme to synthesize DNA sequences, but it was hard to control. The key is to find a way to get the enzyme to add one nucleotide and then stop, so that the sequence can be synthesized one base at a time. Previous approaches tried to obtain that control by using modified nucleotides with a special blocking group that prevents multiple additions at once. After the DNA molecules have been extended by a blocked nucleotide, the blocking groups are removed to allow the next addition.
Previous approaches tried to obtain that control by using modified nucleotides with a special blocking group that prevents multiple additions at once. After the DNA molecules have been extended by a blocked nucleotide, the blocking groups are removed to allow the next addition.
TdT, however, cannot accommodate a blocking group on the nucleotide being added. But Arlow came up with the idea to tether an unblocked nucleotide to TdT, so that after the nucleotide is added, the enzyme remains attached and prevents further additions. After the molecule has been extended, the tether is cut, releasing the enzyme and re-exposing the end for the next addition.
In the first trials, the researchers demonstrated that this technique is not only faster and simpler, but nearly as accurate as other techniques in each step of the synthesis.
“When we analyzed the products using NGS, we were able to determine that about 80 percent of the molecules had the desired 10-base sequence,” Arlow said.
“That means, on average, the yield of each step was around 98 percent, which is not too bad for a first go at this 50-plus-year-old problem. We want to get to 99.9 percent in order to make gene-length DNA.”
Once they can reach 99.9 percent fidelity, they can synthesize a 1,000-base-long molecule with a yield of more than 35 percent, which is currently impossible with existing techniques.
“By directly synthesizing longer DNA molecules, the need to stitch oligonucleotides together and the limitations arising from this tedious process could be reduced,” said Palluk. “Our dream is to directly synthesize gene-length sequences and get them to researchers within few days.”
“Our hope is that the technology will make it easier for bioengineers to more quickly figure out how to biomanufacture useful products, which could lead to more sustainable processes for producing the things that we all depend on in the world, including clothing, fuel and food, in a way that requires less petroleum,” said Arlow.

He added, “Our dream is to make a gene overnight. For companies trying to sustainably biomanufacture useful products, new pharmaceuticals, or tools for more environmentally friendly agriculture, and for JBEI and DOE, where we’re trying to produce fuels and chemicals from biomass, DNA synthesis is a key step. If you speed that up, it could drastically accelerate the whole process of discovery.”
Discuss this and other 3D printing topics at 3DPrintBoard.com or share your thoughts below.
[Source: UC Berkeley / Images: Marilyn Chung, Berkeley Lab]
Stay up-to-date on all the latest news from the 3D printing industry and receive information and offers from third party vendors.
Tagged with: 3d printed dna • dna • dna printing • Joint BioEnergy Institute • Lawrence Berkeley National Laboratory • synthetic dna • uc berkeley • University of California at Berkeley
Please enable JavaScript to view the comments powered by Disqus.
DNA of Things: researchers embed DNA into 3D printed objects » 3D Printing Media Network
(Photo: ETH Zurich | Julian Koch)Stay up to date with everything that is happening in the wonderful world of AM via our LinkedIn community.
A research team based out of ETH Zurich has developed a way to store DNA-like information inside 3D printed objects. The novel approach could allow people to reproduce 3D printed objects without any external digital information, such as original CAD files.
The innovative technology, dubbed the DNA of Things, was pioneered by Robert Grass, a professor in ETH Zurich’s Department of Chemistry and Applied Biosciences, and Yaniv Erlich, an Israeli computer scientist. The pair combined their individual research projects to create the 3D printed, DNA-embedded objects.
As Grass explained: “With this method, we can integrate 3D printing instructions into an object, so that after decades or even centuries, it will be possible to obtain those instructions directly from the object itself. ”
”
As mentioned, the DNA of Things technology draws some work conducted by both researchers. Grass, for his part, developed a method for identifying products with a DNA barcode embedded inside tiny glass beads. The nanobead barcode technology, which has been commercialized by ETH spinoff Haelixa, can be used to identify high-quality food products or to trace geological tests.
Erlich, for his part, pioneered a way to store incredible amounts of data in DNA. The method is theoretically capable of storing 215,000 terabytes of data inside a single gram of DNA. By combining the two inventions, Erlich and Grass have come up with a new type of data storage, one that would allow objects to contain information about their creation.
The white rabbit
To demonstrate the DNA of Things technology, the researchers 3D printed a plastic rabbit that contains roughly 100 kilobytes worth of information about how to 3D print the part. The printing instructions were integrated by embedding tiny DNA-containing glass beads into the printed structure.
The printing instructions were integrated by embedding tiny DNA-containing glass beads into the printed structure.
Using the 3D printed rabbit, the research duo then extracted the information and used it to 3D print a new one. They ended up making five identical 3D printed rabbits based off of the DNA of the original.
“All other known forms of storage have a fixed geometry: a hard drive has to look like a hard drive, a CD like a CD. You can’t change the form without losing information,” Erlich said. “DNA is currently the only data storage medium that can also exist as a liquid, which allows us to insert it into objects of any shape.”
Concealing information
The DNA of Things technology has a number of applications that go beyond simple reproduction. For instance, the research team says it can be used to conceal information inside of everyday objects.
(Photo: ETH Zurich | Jonathan Venetz)The glasses you see in the photo? They contain a short film (1.4 megabytes) about a secret archive from the Warsaw Ghetto in WWII. The information was printed into the lenses. “It would be no problem to take a pair of glasses like this through airport security and thus transport information from one place to another undetected,” Elrich said.
The information was printed into the lenses. “It would be no problem to take a pair of glasses like this through airport security and thus transport information from one place to another undetected,” Elrich said.
The unique technology could also be used to mark and identify things like medication or construction materials. Data about the medication’s quality could be stored directly inside of it, enabling supervisory authorities to conduct quality control tests directly on the product. In construction, the technology could be valuable in telling future construction workers what products were used in a building.
Today, the DNA of Things method is still too costly to be broadly applied, but the research team says that the cost of producing the DNA-embedded objects will decrease (per unit) the larger batches are made—because the bulk of the cost is associated with the DNA synthesis.
Related Articles
Back to top button
3D printing of viable DNA molecules could resurrect the Jurassic era
It is well known that everything is better with lasers, and lasers seem to be able to make DNA 3D printing dangerously close to what we might call "accessible to the masses" , and it's all made possible by Cambrian Genomics, a biotech startup based in San Francisco.
Humans, it looks like we'll have cloned dinosaurs soon. You heard right. Now we can literally print viable strands of DNA. Of course, all this is not yet so close to real life, but these developments are clearly the first step into the real world of Jurassic Park. In a video published on Bloomberg, Cambrian Genomics co-founder Heinz Austin even claims that we will soon have cloned dinosaurs:
“This technology will allow us to actually bring back dinosaurs, bring back extinct animal species, and in addition, create new life forms that will help us survive in space and go to other planets”
Laboratory and synthesis of DNA molecules at 3D printing help
And apparently we will get not just dinosaurs, but dinosaurs capable of living in space. Of course, for a couple more years we may not worry about our laboratory-grown space tyrannosaurs, with laser blasters on their backs and devouring people, but today it is already becoming just a matter of time. Anyone can create super space dinosaurs and they'll be great.
Anyone can create super space dinosaurs and they'll be great.
The cost of printing various genomes and their components, from left to right: protein, plasmid, bacterial genome, yeast genome, human genome.
Amazingly, it turns out that DNA printing is not a new technology at all. We have been doing this for many years now. This technology is used to create drugs, to study dangerous pathogens, and we can even program DNA to store huge amounts of data. The catch is not the ability to synthesize DNA, but the cost of the process. Printing the human genome using traditional methods would cost billions of dollars, and in doing so, we would get only one set of human chromosomes. This high price is due to the high failure rate in DNA cultivation. An incredibly large amount of DNA can be grown very quickly, but a large percentage of them will contain errors and defects that make them useless and therefore need to be sorted. As you can probably guess, sorting billions of strands of DNA is a bit of a time consuming process.
But the new technology developed by Cambrian Genome speeds up this process many, many times over thanks to laser printers for DNA sorting, which makes this process much faster, and therefore more affordable. How accessible will it become? A DNA laser printer will produce more viable DNA at one time than all the machines in the world can produce in one year. Turning the DNA printing process into something that can be accessed by more than just some multi-billionaire or super villain who wants to breed a cosmic thug dinosaur would probably serve humanity well.
Growing synthetic DNA is not the easiest process, but, in fact, they are grown in a sense, like grass. Millions of strands are formed by mixing basic DNA compounds in a specific sequence. After the filaments are formed, they are detached from the structures on which they have grown and then attached to a one micron ball. After one strand of DNA is attached to each bead, the strand itself is cloned in such a way that each bead has multiple copies of the strand that was attached to it first. The beads are then moved onto a small glass slide and a laser printer examines the DNA strands and selects those beads that contain the correct strands. Thus, full-fledged DNA molecules are sent to special cells, and all unwanted or defective DNA remains on the glass.
The beads are then moved onto a small glass slide and a laser printer examines the DNA strands and selects those beads that contain the correct strands. Thus, full-fledged DNA molecules are sent to special cells, and all unwanted or defective DNA remains on the glass.
Genome lab 3D laser printer
This process starts with one million strands of DNA grown in the lab and ends up with a billion cloned, quality strands sorted at up to a hundred strands per second. As Heinz put it, this will make DNA a commodity.
"Anyone can be a genetic designer"
Of course, that's a little scary considering some of the consumer products that have managed to hit the market today, but it's surprising. We are already living in the future. We are no longer only printing cases for our smartphones, we are able to create life through 3D printing. Yes, it will probably be 3D printed life that will turn into space dinosaurs that can eat us, but, you must admit, this is a truly exciting prospect.
If you want to learn more about the DNA laser printing process, check out this video and slideshow that explains the process in more detail:
Article prepared for 3DToday.ru print
Swiss and Israeli researchers proposed to introduce DNA into objects with encoded instructions for their production at the production stage. As an example, they 3D printed a model of a rabbit, encoded a print file in it, and then successfully restored it. Article published at Nature Biotechnology .
There are many projects designed to preserve the key elements of human civilization in the event of a global catastrophe. For example, in Svalbard, since 2008, there has been a secure repository for seeds of the main agricultural plants, and in 2020, a repository for all open source programs published on GitHub will be created near it. In addition, even though there are no global catastrophes, people often face the problem of recreating technologies that were used several decades ago. For example, a significant part of the documentation of the most powerful rocket in the history of Saturn V was lost in the first decades after the end of the program.
For example, a significant part of the documentation of the most powerful rocket in the history of Saturn V was lost in the first decades after the end of the program.
A group of scientists led by Robert Grass from the ETH Zurich and Yaniv Erlich from the Erlich Lab proposed saving drawings and other documentation to recreate objects and technologies directly in them. They chose DNA as a carrier, which under certain conditions can provide an extremely high density of information storage, as well as a duration of thousands of years. The method of recording in DNA was developed by Erlich and colleagues, and is based on redundant "fountain" coding, which allows you to completely restore information even if a significant part of the material is lost. We talked more about coding with this method in 2017.
As the authors applied the DNA writing approach to 3D printing plastic, they used a method previously developed by the group led by Grass to embed DNA molecules into amorphous silicon particles that isolate DNA from external influences. In addition to protection, this makes it possible to introduce DNA into filaments for 3D printers that have different compositions, and also not affect the properties of the printed object.
In addition to protection, this makes it possible to introduce DNA into filaments for 3D printers that have different compositions, and also not affect the properties of the printed object.
Researchers have experimentally shown a complete cycle of encoding and decoding information using the example of a Stanford rabbit figurine, a standard 3D model used in scientific works from various fields. After creating an STL file with a 3D model, they synthesized 12,000 oligonucleotides (short fragments) of DNA with a 5.2-fold redundancy. This means that even if 80 percent of the oligonucleotides are lost, the original file can be restored from the remaining amount.
After synthesis, they embedded DNA into silicon spheres, mixed it with polycaprolactone, and formed a filament thread from it for a 3D printer. The researchers then printed a model of the rabbit and cut off a small part of it for restoration. At the last stage, they were able to isolate oligonucleotides from the material and, using the PCR method, completely restore the encrypted file, despite the loss of 5.